Next Gen LSDA
Data Submission Guidelines
Guidelines
The guidelines presented here intend to provide data submission requirements and recommendations regarding overall information organization, file naming and file structures, for use by investigators archiving data with NASA’s Life Sciences Data Archive (LSDA). These guidelines have been developed to enable consistent, high quality, computable data. While some guidelines are not requirements, they are highly recommended to ensure data from NASA-funded studies are findable, accessible, interoperable, and reusable.
File Organization
The directory structure of your submittal can significantly contribute to understanding the data package and efficient consumption of the data. File organization should move from general information in the top directories to increasingly specific information in the sub-directories. This section outlines a number of recommendations to support the development of a well-formed data package.
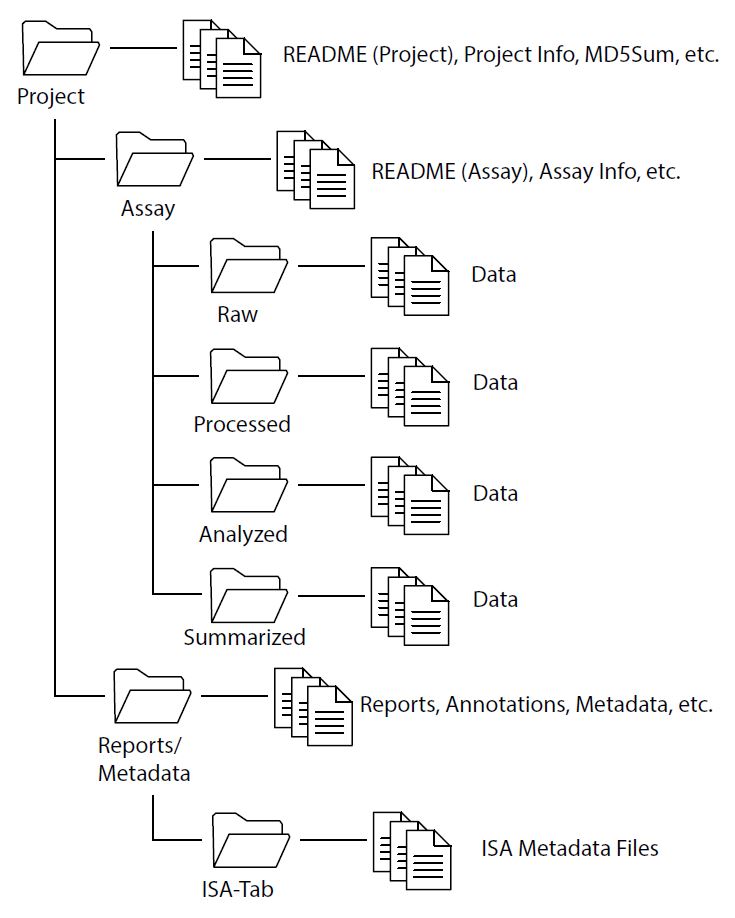
Organize files into a logical directory structure, and not as a loose set of files within a single folder.
- Studies typically follow a progression of data evaluation from raw to summarized findings.
- Group files based on Project, Assay (or data type), and ancillary or metadata. (see Figure 1)
The primary level directory folder should contain:
- A README file describing the Project and dataset, the various directories and files, and a description of the relationship of the files.
- See README File Guidelines for file best practices.
- A data manifest of all the files in the complete data package, as a reference to ensure successful data submission.
The data submission should contain a single manifest of all files in the submission, to include the relative
file path name starting from the main data directory (do not include the full local file path)
- The data manifest should contain two columns separated by a tab:
- Relative file path and name, relative to the main folder
- File size in bytes
file_location size_bytes /project/assay/raw/missionX_subjectA_assay.tar 3139235840 /project/assay/raw/missionX_subjectB_assay.tar 2713293944
- The manifest may be generated using simple scripts, once the file organization is complete
and include the relative file path of the data directory. Do not include local directories names
that will not be part of the submission package. Below are examples scripts of how to generate the
manifest, based on operating system. These are just a starting point to serve as an example and help the data submitter get started.
- Unix/Linux/Mac/Gnu>$ find /data/directory/project/ -printf ‘%p\t%s\n’ > /data/directory/project/manifest.txt
- WindowsPS > $StartDir=”\data\directory\”
PS > Get-ChildItem -Path $startDir -File -Recursive | Select-Object @}, @} | Export-Csv ‘$manifest.txt’ -NoType -Delimiter “`t”
- Unix/Linux/Mac/Gnu
- The data manifest should contain two columns separated by a tab:
- An MD5 Checksum is required for each data and metadata file, but may also be provided for every file,
including README and annotation files (see MD5 Checksum Section). It is common to include a single md5sum
file in the top level directory, but multi-part md5sum files may be provided for each sub-directory.
Below is a common example of the contents of an md5sum file.
md5sum file_location 957c168884ccc1dbfb0e1028ffd1e53e /project/assay/raw/missionX_subjectA_assay.tar af1b39494c2c66a21add7e80a3c5d7d3 /project/assay/raw/missionX_subjectB_assay.tar
Secondary/tertiary level directory folders should contain:
- Files representing different levels of abstraction grouped within their own folder.
- Folders with data should only contain files of the same type. Do not intermix different file types within the same folder (e.g. all files related to an RNASeq assay should be contained within a single directory structure).
- Nested folders of related data should contain README files to more completely describe the structure of each file type.
- Data integrity checks (MD5 Checksum) files may be included at each directory level.
Additional files – such as summary findings, reports and statistics – may be included within the primary or secondary level directory folders, but not intermixed with data files. A sample directory structure is provided in Figure 1:
File Naming Convention
Descriptive file names are an important aspect of organizing, sharing, and managing data files. Develop a naming convention based on elements important to the project. Additionally, do not reuse simple file names multiple times in different directories. Ideally, use file names that are unique to the data set. This section builds on the guidelines of the Princeton University Library best practices for file naming. The most important aspects to remember about file naming are to be consistent and descriptive in naming and organizing your files so that it’s obvious where to find a file and what it contains
- Excel spreadsheet example:
- <grant#>_<experiment-id>_<desc>.xslx
- MRI images taken in different positions:
- <university>_<study>_<type>_<position>_<yyyymmdd>.dcm
- Human genome sequence:
- <institution>_<platform>_<mission>_<subjectID>_<sequenceNumber>_<yyyymmdd>.fastq
- <institution>_<platform>_<mission>_<subjectID>_<yyyymmdd>.bam
- <institution>_<platform>_<mission>_<subjectID>_<VCFtype>_<yyyymmdd>.vcf
Metadata Guidelines
- Create one README file for each data type whenever possible. It is also appropriate to describe a "dataset" that has multiple, related, identically formatted files, or files that are logically grouped together for use (e.g. a collection of Matlab scripts). When appropriate, also describe the file structure that holds the related data files.
- Name the README file so that it is easily associated with the data file(s) it describes.
- Write your README document as a plain text file, avoiding proprietary formats (such as MS Word) whenever possible. Format the README document so it is easy to understand (e.g. separate important pieces of information with blank lines, rather than having all the information in one long paragraph).
- Format multiple README files identically. Present the information in the same order, using the same terminology.
- Use standardized date formats. Recommended format: W3C/ISO 8601 date standard, which specifies the international standard notation of YYYYMMDD or YYYYMMDDThhmmss.
- Follow the scientific conventions for your discipline for taxonomic, geospatial and geologic names, and keywords. Whenever possible, use terms from standardized taxonomies and vocabularies
README File Contents — Description (Minimal recommendations in bold)
- Provide a title for the dataset
- Name/institution/address/email information for
- Principal investigator (or person responsible for collecting the data)
- Associate or co-investigators
- Contact person for questions
- Date of data collection (can be a single date, or a range)
- GMT Standard (not local time zone)
- Information about geographic location of data collection
- Keywords used to describe the data topic
- Language information
- Information about funding sources that supported the collection of the data * Minimal recommendations in bold
File Contents — Data and File Overview (Minimal recommendations in bold)
- For each filename, a short description of what data it contains
- Format of the file if not obvious from the file name
- If the data set includes multiple files that relate to one another, the relationship between the files or a description of the file structure that holds them (possible terminology might include "dataset" or "study" or "data package")
- Date that the file was created
- Date(s) that the file(s) was updated (versioned) and the nature of the update(s), if applicable
- Information about related data collected but that is not in the described dataset
README File Contents — Sharing and Accessing Information (Minimal recommendations in bold)
- Restrictions placed on the data
- Links to publications that cite or use the data
- Links to other publicly accessible locations of the data
- Recommended citation for the data, especially if some parts of the data persist in a data service other than the LSDA
README File Contents — Methodology (Minimal recommendations in bold)
- Description of methods for data collection or generation (include links or references to publications or other documentation containing experimental design or protocols used)
- Description of methods used for data processing (describe how the data were generated from the raw or collected data)
- Any instrument-specific information needed to understand or interpret the data
- Standards and calibration information, if appropriate
- Describe any quality-assurance procedures performed on the data
- Definitions of codes or symbols used to note or characterize low quality, questionable info or outliers people should be aware of
- Point(s) of contact for sample collection, processing, analysis and/or submission
README File Contents — Data Specific Information (Minimal recommendations in bold)
- Count of number of variables, and number of cases or rows
- Variable list, including full names and definitions of column headings for tabular data (and spell out abbreviated words)
- Units of measurement
- Definitions for codes or symbols used to record missing data
- Specialized formats or other abbreviations used
README File Contents — Examples of Metadata Standards: Table 1
| Source | Content |
|---|---|
| BioPortal | Biomedical ontologies – comprehensive resource for molecular, biological, and medical ontologies |
| Integrated Taxonomic Information System | taxonomic information on plants, animals, fungi, microbes |
| NASA Thesaurus | engineering, physics, astronomy, astrophysics, planetary science, Earth sciences, biological sciences |
| GCMD Keywords | Earth & climate sciences, instruments, sensors, services, data centers, etc. |
| USGS Thesaurus | agriculture, forest, fisheries, Earth sciences, life sciences, engineering, planetary sciences, social sciences etc. |
| Getty Research Institute Vocabularies | geographic names, art & architecture, cultural objects, artist names |
LSDA is transitioning to an Investigation-Study-Assay (ISA) framework for metadata which will provide detailed context and descriptions of experiments within the archive. These metadata enhance data discoverability within the archive database, and ensure the data can be efficiently exchanged and integrated for use by future research.2 Specifications for submitting data in compliance with ISA documentation will be forthcoming
For now, please see the ISA documentation for an introduction to the topic: https://isa-specs.readthedocs.io/en/latest/isamodel.html
Data File Formatting Guidelines
This section will address the requirements and recommendations for the structure and description of the two most common forms of data:
- User created columnar / tabular data.
- Spreadsheets and text data files.
- Encoded data.
- Unique file format based on a specification.
- Commonly found in images and vendor specific file formats.
For each form of data, there will be a limited set of Requirements, meaning data that does not adhere to these characteristics
will not be acceptable to the LSDA; and a set of Recommendations, which are highly desired characteristics, but not required.
The most common form of a data file is a tabular data frame, like a spreadsheet.
The table form consists of rows and columns of information.
Each column contains information regarding a specific attribute, and each row consists
of various data related to an entity or observation.
The columns have headers that describe the data in each column should represent a specific variable
with a well described information type and structure.
At a minimum, the rows should have a unique index, made up of one or more columns.
In general, data files should be machine-readable first and human-readable second.
The data should be developed for pulling the information into analytical systems
(such as Jupyter Notebooks, Matlab, etc.) and not for presentation purposes.
Examples: tab or comma delimited text files, Microsoft Office Excel spreadsheets, SAS/STAT data, etc.
- Each data file(or worksheet within an Excel workbook) should only have one type of data per file.
- A data file must be an individual file (or Worksheet within an Excel Workbook).
- A Workbook may consist of multiple Worksheets, with each Worksheet treated as a unique data file.
- Do not include data from multiple assay types in a single file or worksheet, and do not include statistics, summary data, or graphs in the data file.
- The primary exception to this rule is the document header information, which should be limited to data definitions or description of data issues.
- Excel spreadsheets should not include document header information. All non-data annotations present in a spreadsheet should be included in a separate, non-data Workbook, Worksheet, or text file
- Do not intermix raw, analyzed, and summarized data within a single file.
- A data file must be an individual file (or Worksheet within an Excel Workbook).
- Each data file (or worksheet within an Excel workbook) should include only a single data table.
- Do not stack tables, such that there are multiple data sets within a given column.
- Do not arrange tables alongside one another, such that there are multiple tables in a given row.
- Columnar data must consist of a header row that describes each column.
- Data files without a descriptive row will not be accepted.
- More than one data table header row will not be allowed.
- A header row, describing each column, should not be confused with the document header commentary
- Data definitions must accompany each unique type of data. The definition must explain the data contained in each column and include the actual header row name for each column.
- File structure and data value descriptors may be included within the document header area - see “Columnar/Tabular Data Recommendations”..
- If the definitions are not included within the document header, the definitions should be provided in a separate Data Dictionary document.
- Where the data file is based on an open specification, a link to the specification may be provided – within document header, README, metadata documentation, etc.
- Adherence to Tidy Data guidelines for data structure is recommended, in which:
- Columns represent variables
- Rows represent observations
- Tables consist of observational units
- Individual text files are preferred over Excel Workbooks.
- Tab delimited columns is preferred over comma delimited, as commas are often included within single data fields
- Multiple data files of single data sets are better than large single files representing many data sets.
- Descriptive document header information may be included within the file, before the start of the data, which is common in open data specifications.
- Document header information consists of instructions, metadata, and/or annotations to inform the use of the data.
- The term “document header” implies it occurs prior to the data. All comments and annotations should be placed before any data and not be intermixed with data rows.
- Document header information should be identified by a non-data character as the first character in the line (i.e. “#” or “##” will indicate the line represents a comment or metadata).
- Example:
# This is a comment line and may include non-data information
# Comment lines must occur before the first line of data - If metadata elements, column descriptors, or other reference terms are included in the document header, these elements should adhere to a standard format to automate information extraction.
- Data annotations may be included, such as conditions on certain information, file specification links, or other information pertinent to the data file.
- Example:
Encoded files consist of data that adhere to a specific encoding and format that must be read
using a specific software or must be decoded using a file format specification.
Many reasons exist to encode a file. Most software vendors employ unique file encodings
to ensure compatibility with and use of their software.
Additionally, data may be encoded for the purpose of addressing specific data needs,
obscuring information, and reducing file sizes, to name a few.
Image files represent a common type of file based on a file encoding.
In order to use encoded data, a data specification must be published so that
consumers of the data will know how to decode the data. In the case of common
files such as images, file reader tools should be available so that data users
are not required to develop their own software. In the case of text encoded data,
these data should adhere to common standards, such as base64 encoding, which can
easily be handled within most programming languages.
Examples of commonly used file specifications:
Open and Proprietary Encoded Data Requirements
- Encoded data files must adhere to an open file standard, except for the following cases:
- The non-open standard is very commonly used by the research community, and tools are readily available to read or transform the data.
- Example – MS Office documents, standard image formats, etc.
- There are no open alternatives to the represented data.
- The non-open standard is very commonly used by the research community, and tools are readily available to read or transform the data.
- A description of each type of encoded data must be provided, whether in a README file or other annotation file.
- A separate descriptive document must be provided, even if annotations are included within the document header.
- For data descriptions provided within a README file:
- An external file specification may be referenced, preferably within the README file.
- For multiple data types and data specific file encodings, each data type must be described in an annotation file, such as a README file.
- Attributable data, such as Personally Identifiable Information or Personal Health Information, MUST be removed from or obfuscated within the encoded file.
- Common examples are medical images (DICOM, pathology) and DNA/RNA BAM files.
- Encoded files should reside within their own directory folder, separate from text files.
- Encoded files should reside within their own directory folder, separate from text files.
- Data specific README files are encouraged.
- A description of relevant field tags encoded within each data type is very helpful to data consumers.
Data Integrity and Quality
Data integrity is of utmost concern to the NASA Life Sciences Data Archive.
The first line of data integrity verification is based on a handshake between the data submitters and the LSDA team.
In order to ensure research data are accurately transmitted upon submission to the LSDA, verification of a cryptographic hash will be employed.
Specifically, the LSDA will require those submitting data to include an MD5 (128 bit) hash for each file.
The specification for the Message-Digest Algorithm 5 (MD5) can be found here: https://tools.ietf.org/html/rfc1321.
An MD5 Checksum is required for each data and metadata file, but may also be provided for every file,
including README and annotation files (see MD5 Checksum Section).
It is common to include a single md5sum file in the top level directory, but multi-part md5sum files may be provided for each sub-directory.
Below is a common example of the contents of an md5sum file.
Example contents of md5sum file:
| md5sum | file_location |
|---|---|
| 957c168884ccc1dbfb0e1028ffd1e53e | /project/assay/raw/missionX_subjectA_assay.tar |
| af1b39494c2c66a21add7e80a3c5d7d3 | /project/assay/raw/missionX_subjectB_assay.tar |
DO NOT encrypt files directly. Encrypting specific files requires the use of passcodes, encryption keys, or certificates. The LSDA cannot accept and manage passwords and keys for individual files. Encrypted data will be automatically rejected and the submitter will be required to submit data that is not encrypted. Encryption of sensitive data shall be accomplished by transmitting the data using a secure method (i.e. SSL, HTTPS, SFTP, etc.) and by relying upon the encryption capability built into the computer’s operating system or underlying hardware.